July 28, 2024, by Brigitte Nerlich
The genome as autoencoder: A new biological metaphor
I am just back from a walk thinking about Kevin Mitchell and Nick Cheney’s recent paper (preprint) on the genome as autoencoder, rather than a blueprint or recipe. This paper caused quite a stir and you can find a good summary in this post by Jessica Hamzelou for the MIT Technology Review.
Walking along, I was wondering whether there was anybody out there who understood metaphors, genomics and machine learning/AI to discuss this paper with me, especially the issue of lay and expert understanding of these metaphors… and suddenly Claude, the Anthropic chatbot, popped into my head.
So, on my return, I had a little chat with Claude about the paper. I summarised the gist of the paper and Claude thought that “using an auto-encoder as a metaphor for the genome is intriguing”. It then explained to me a little bit more what an autoencoder was and how it could be used to shed light on aspects of the genome.
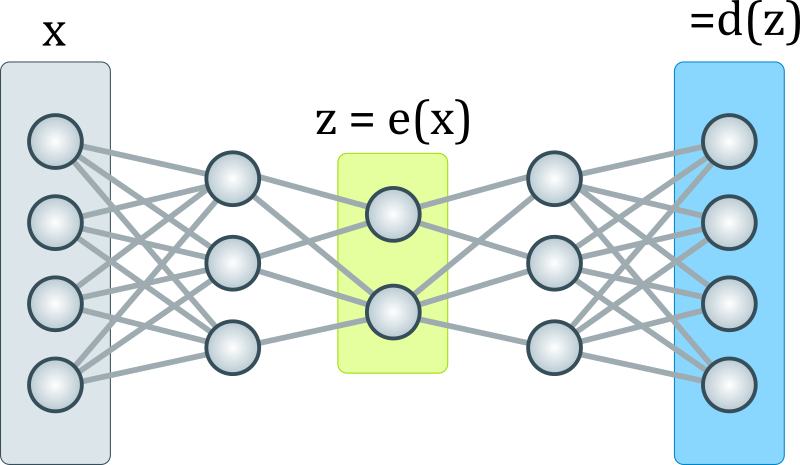
The genome as autoencoder
“An auto-encoder is a type of neural network that learns to encode data into a compressed representation and then decode it back to the original input. This metaphor could potentially capture several important aspects of genomics:
a) Compression: The genome contains a vast amount of information in a relatively compact form, similar to how an auto-encoder compresses data.
b) Generative capability: Auto-encoders can generate new data similar to the input data, which could be analogous to how the genome guides the development of an organism.
c) Complex interactions: The hidden layers in an auto-encoder could represent the complex interactions between genes and environmental factors.
d) Robustness: Auto-encoders can often reconstruct input from partial or noisy data, which might be similar to how organisms can develop normally despite some genetic or environmental perturbations.”
Pros and cons of the metaphor
Claude generally thought that this metaphor addresses some of the shortcomings of previous metaphors, such as the blueprint and the recipe and “better captures the idea of the genome as a complex, interactive system rather than a simple one-to-one mapping or linear process.”
However, Claude also pointed out that, as with all metaphors, this new metaphor has some limitations:
- “Auto-encoders are typically trained on static data, while genomes have evolved over millions of years.
- The genome interacts with environmental factors in ways that might not be easily captured by the auto-encoder metaphor.
- The metaphor might not adequately represent epigenetic factors or the role of non-coding DNA.”
(For those who want a more technical summary of the autoencoder/genome metaphor mapping, see the addendum below which Claude generated after I gave it the abstract of the paper)
The heuristic value of the metaphor
Claude also came up with a good idea for future research, namely the need to evaluate what one might call the heuristic value of the metaphor for genomic research: “To fully evaluate the effectiveness of this metaphor, we’d need to consider how well it helps researchers and students understand genomic concepts, and whether it leads to new insights or research directions.”
After that I said that this was all great, but lay people who don’t have background knowledge in machine learning and AI (unlike about blueprints and recipes) would have difficulties using this metaphor to understand how genomics works, and I asked what one could do about it. So, Claude came up with more user-friendly metaphors capturing aspects of the autoencoder metaphor.
Proposals for accessible metaphors
(Some of these metaphors have been discussed in the literature, especially the music and theatre metaphors for example here, and here, and here and here)
- “The Compression and Decompression Analogy: Imagine the genome as a highly sophisticated zip file. When you compress a file, you’re not storing every single detail, but rather a compact representation that can be used to recreate the original. Similarly, the genome doesn’t contain a full description of the organism, but a compressed set of instructions that can “unfold” into a complete organism.
- The Recipe Book with Flexible Ingredients: Instead of a single recipe, think of the genome as a magical recipe book. This book doesn’t just list ingredients and steps, but describes relationships between ingredients. As you “cook” (develop), the recipes adapt based on what’s available in your kitchen (environment). The final dish (organism) emerges from these flexible, interacting recipes rather than from following a rigid set of instructions.
- The Musical Score Analogy: Consider the genome as a complex musical score. The notes on the page don’t directly represent the music you hear. Instead, they provide a set of instructions that, when interpreted by musicians (cellular processes), produce the final symphony (the organism). Different performances (individuals) might have slight variations, but they’re recognizably the same piece.
- The Seed and Garden Metaphor: Think of the genome as a seed, and the process of development as growing a garden. The seed doesn’t contain miniature plants, but rather the potential to grow into plants. The final form of the garden depends on the interaction between the seed’s instructions and the environment (soil, water, sunlight). This metaphor captures the idea of latent potential and environmental interaction.
- The Improv Theater Script: Imagine the genome as a script for an improv theater performance. It doesn’t dictate every word and action, but provides characters, relationships, and key plot points. The final performance (the organism) emerges from the interaction between these guidelines and the actors’ choices (cellular processes and environmental factors).
These metaphors, while still imperfect, attempt to capture key aspects of the auto-encoder metaphor in more accessible terms:
- The indirect nature of genomic encoding
- The emergence of complex traits from simpler instructions
- The interaction between genetic information and environmental factors
- The potential for variation within a consistent overall pattern”
Implications for public understanding and science communication
“By using these more accessible metaphors, we can help lay people understand that:
- Genes don’t directly specify traits, but rather provide a framework for development.
- The same genetic information can lead to different outcomes based on environment and chance.
- Complex traits emerge from the interaction of many genes, not from single genes acting in isolation.
- There’s both consistency and flexibility in how genetic information guides development. [italics added]
These concepts align with the core insights of the auto-encoder metaphor while being more accessible to those without a background in machine learning.”
After I thanked Claude for these useful proposals, it came up with a bonus proposal, so to speak, relating to science communication and tuning metaphors to audiences:
“I’m glad you found these alternative metaphors useful. They aim to bridge the gap between the technical precision of the auto-encoder metaphor and the need for accessibility in explaining complex genomic concepts to a broader audience.
These kinds of accessible explanations are crucial for public understanding of science, especially in fields like genomics that have significant societal implications. They can help people grasp important concepts like genetic complexity, gene-environment interactions, and the probabilistic nature of genetic influences on traits. [italics added]
If you’re working on communicating these ideas to a non-specialist audience, you might consider using a combination of these metaphors. Different analogies might resonate better with different people, or might be more suitable for explaining particular aspects of genomic function.”
What did I learn?
I found this chat with Claude really enlightening. It gave me lots of insights not only into the meaning of the autoencoder metaphor which, for somebody like me without any understanding of machine learning, was quite obscure in places, but also into how to dispel that obscurity with other more accessible metaphors.
Claude also stressed that metaphors must be attuned to audiences. For metaphors to work we need some knowledge of the source domain (e.g. of autoencoders) and the target domain (of genomics) and for metaphors to work in science communication we also need to have some knowledge of the knowledge or lack of knowlege that audience bring to the understanding of metaphors. Only then can we ensure, at least a little bit, that the stories we tell about science resonate with peoples experience, knowledge and values.
And finally, it was refreshing to read Claude extolling the virtues of a dynamic/process rather than static/product image of genomes, development and organisms. Some think that this view of genomics has as not yet penetrated the public sphere. But I wonder…. if Claude picks this up, it must be out there….
Image: Openclipart
Addendum
The auto-encoder metaphor – a more detailed dissection
- “Genome as a generative model: The core of Mitchell and Cheney’s proposal is that the genome encodes a generative model of the organism, analogous to a variational autoencoder (VAE). This is a more sophisticated version of the auto-encoder metaphor we discussed earlier.
- Latent variable space: Mitchell and Cheney suggest that the genome comprises a compressed space of latent variables, represented by DNA sequences. This is directly analogous to the compressed representation in the hidden layer of an autoencoder or VAE.
- Decoding through development: The idea that these latent variables are ‘decoded’ through developmental processes aligns well with the decoding phase of an autoencoder.
- Connectionist network: Mitchell and Cheney describe the genome as a connectionist network, with weights encoded by evolution. This draws a parallel between genomic evolution and the training process of neural networks.
- Energy landscape and Waddington’s epigenetic landscape: The concept of the latent variables shaping an energy landscape that constrains development is a powerful extension of the autoencoder metaphor. It provides a link to Waddington’s classic concept of the epigenetic landscape.
- Emergent properties: Mitchell and Cheney argue that this model accounts for complex genetic architecture, robustness, and evolvability. These emergent properties are indeed characteristics of deep learning models like autoencoders.
- Multiplexed disentangled representations: This concept from artificial and neural systems is applied to explain the independent selectability of traits, which is an interesting extension of the metaphor.”
ADDENDUM 15 September 2024: Wow, now there is an AI generated podcast about this paper!

Here are a few random comments from a friend who read the blog. (I have transcribed them here as faithfully as I could, but there might be some errors!) So these notes are based on a ‘chat’ with a ‘real person’ based on a chat with an artificial person! (The real person is not a ‘real’ expert in ML or AI)
First of all, I should stress that one should not confound what Claude said about autoencoders with what the article says. That would need some more disentanglement.
As @cj_baron pointed out on, normally metaphors map what’s familiar onto something unfamiliar, e.g. recipe or book onto genome. Autoencoder is familiar to some but probably not to the majority of people or ‘publics’.
We need to distinguish between metaphors for the genome, such as blueprint, and metaphors for evolution and development, such as autoencoders.
There is also another aspect that might need disentangling and that is whether we are saying that autoencoders turn inputs into hidden layers and then into outputs, rather as evolutionary history is represented in genomes which then become phenotypes. That is a bold philosophical statement, not just a metaphor.
Setting all that aside, if we stick to genes and genomes and the autoencoder metaphor, my friend said that it might be wrong to say that genes are a compressed version of anything. They are an efficient and compact code (up to a point) but also contain a lot of redundancy and waste.
So, a better metaphor for the conversion of an input to a sparse representation and back into an output might be that of ‘back translation’, i.e. where content is translated to a new language and then translated back to its original language for comparison to the source text. This is not about creating new data.
Hidden layers: Think of a letter you receive from Auntie Flossy. You read the letter and you make some notes on a piece of paper on what to say in your reply. The next morning you compose your response to her letter not by looking at the letter again but at your notes. The notes are the hidden layers, and they never appear to the outside world as part of the input or the output.
Complex interactions: The hidden layers in an auto-encoder are like the stages in a Chinese whispers game. They are not in direct interaction with the outside world.
Autoencoders do not use single units to represent tokens or features. They are more like a text that uses a ‘word cloud’ to represent an abstraction like ‘freedom’. And it may use many of the same words that occur in discourse about a wholly different concept.
As for the musical score metaphor. One can perhaps regard the score as a target… A musical score does not represent the resulting music exactly, but rather the ‘target’ towards which the performer is aiming, albeit imperfectly.
The actions of the genome can perhaps be compared to ‘rules of engagement’. The rules do not dictate every single action taken but the parameters within which actions are taken.
Discuss!!
After Kevin Mitchell published a thread on Bluesky on a peer-reviewed version of the paper on 18 January, I gave Claude the words, not pictures, of the thread and asked again to tell me what it’s all about. Again, I found the answer quite useful. I wonder what others think:
Claude:
Yes, I can help break down this fascinating paper’s main argument. The authors are proposing a new way to conceptualize how genomes encode organisms, using analogies from machine learning – specifically variational autoencoders (VAEs) and dynamical systems.
Here are the key points:
Instead of thinking of the genome as a blueprint or recipe, they suggest viewing it as a compressed representation of an organism’s form, similar to how VAEs learn compressed representations of data.
Gene regulatory networks act like the decoder part of a VAE, “unpacking” this compressed information during development. But crucially, this decoding process works through physical and chemical dynamics, not like a computer program executing instructions.
The famous “Waddington’s landscape” concept in developmental biology (which shows how cells develop along certain preferred pathways) can be understood mathematically as emerging from these dynamical networks.
This model helps explain several puzzling features of biology:
Why most genetic variants affect multiple traits (pleiotropy)
Why most traits are affected by many genes (polygenicity)
How organisms maintain stability despite genetic and environmental variation
How evolution can still select for specific traits despite this complexity
They propose that evolution acts like a learning algorithm, optimizing these compressed representations over time to produce reliable development of species-typical traits.
The authors acknowledge they’re using a technological metaphor (machine learning) to understand biology, which can be dangerous if taken too literally. However, they argue this conceptual framework captures important features of biological reality better than previous metaphors like “blueprint” or “recipe.”
… I [Claude] find the connection between VAEs and biological development particularly interesting, as it suggests a mathematical framework for understanding how complex organisms can develop reliably from compressed genetic information.