
February 22, 2019, by Brigitte Nerlich
The exposome – the what?
I recently came across the term ‘exposome’ (roughly, the sum total of everything we are exposed to) and started to read up on it. But I just don’t know what to make of it… is it merely humbug or is there more to it? In this post I want to summarise a few milestones in the emergence of this new field and then ask you readers what you make of it. (This is in the context of my interests in genomics, synthetic biology, epigenetics, microbiomics and so on…)
Creation of a new field
Interestingly, one can date the birth of the ‘exposome’ quite exactly to 2005. This was the year that Christopher Paul Wild published an editorial in the journal Cancer Epidemiology, Biomarkers and Prevention entitled “Complementing the Genome with an ‘Exposome’: The Outstanding Challenge of Environmental Exposure Measurement in Molecular Epidemiology”. If one wanted to be facetious, one could say that the exposome is ‘complementary genomics’…. I won’t do that. Less facetiously, one could say that this conceptual division resurrects or at least mirrors the nature/nurture dichotomy.
The article appeared two years after the completion of the Human Genome Project which revealed just how much we don’t know about genetics and genomics. It laid the foundations for digging deeper into how proteins work (proteomics), how transcription factors work (transcriptomics), how metabolites work (metabolomics)…. and of course, how gene-environment interactions work. As Wild pointed out: “The sequencing and mapping of the human genome provides a foundation for the elucidation of gene expression and protein function, and the identification of the biochemical pathways implicated in the natural history of chronic diseases, including cancer, diabetes, and vascular and neurodegenerative diseases. This knowledge may consequently offer opportunities for a more effective treatment and improved patient management.”
There was talk about precision medicine, but hopes invested here have fallen short of the visions they conjured up. This may partly be due to lack of understanding of gene-environment interactions, most importantly the interaction between genes (metabolites, transcription factors….) and environmental exposures.
This seems to be where the exposome comes in. As Wild says: “There is a desperate need to develop methods with the same precision for an individual’s environmental exposure as we have for the individual’s genome. I would like to suggest that there is need for an ‘exposome’ to match the ‘genome.’ This concept of an exposome may be useful in drawing attention to the need for methodologic developments in exposure assessment.”
Exposome research would be closely linked to epidemiology, environmental health, toxicology and human exposure assessment. Epidemiologists would work with new ‘technologies’ developed in the context of proteomics and metabolomics, as well as with new types of researchers, such as “biostatisticians, experts in bioinformatics, and laboratory and environmental scientists”.
A field begins to flourish
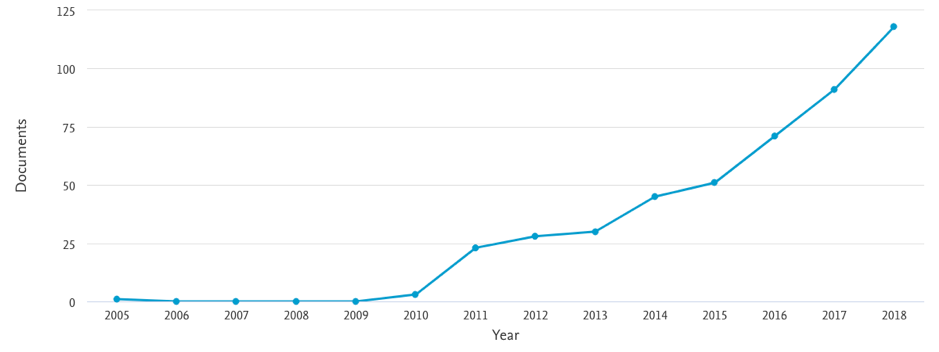
The concept of the exposome lay fallow for a while before beginning to be used more and more frequently around 2011. Overall numbers of publications on the exposome are, however, still quite modest (see figure 1, derived from Scopus)
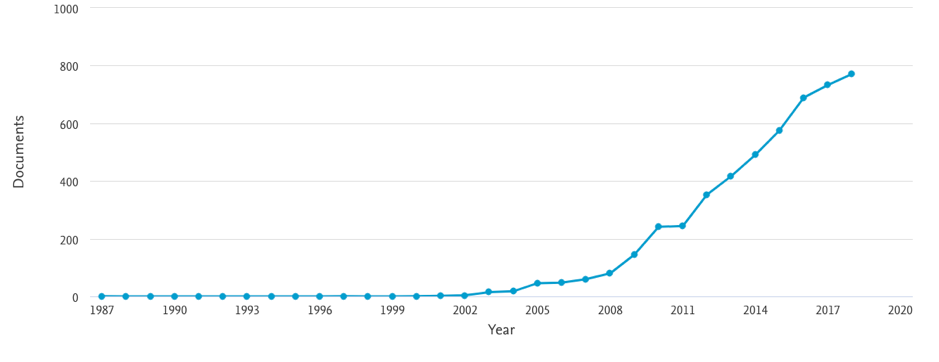
That was also the time when epigenetics accelerated its popularisation and when, indeed, some researchers began to establish links between the epigenome and the exposome (see figure 2). (I’ll leave aside here links between the exposome and the microbiome…).
The epigenome ”is made up of chemical compounds and proteins that can attach to DNA and direct such actions as turning genes on or off, controlling the production of proteins in particular cells” and so on. The action of the epigenome may be influenced by what goes on inside, but also outside the human body – in the environment. That’s where the exposome comes in.
In addition to epigenetics overall beginning to flourish, a subfield of epigenetics also began to garner attention at that time, namely environmental epigenetics, which, given its focus on the environment, is a bridge between epigenetics and exposome research or what some call exposomics.
In 2012, Wild published a follow-up article in the journal International Journal of Epidemiology, entitled “The exposome: from concept to utility”. Between 2005 and 2011 there were advancements not only in ‘omics’ ‘technologies’, but also other technologies that now could be used to monitor environmental exposures more systematically. As Wild says: “Technological advances in biomarkers, personal monitors, imaging etc., offer ways to construct the exposome with increasing completeness.“ As with genome research accelerating after the invention of new sequencing technologies, so exposome research was pushed forward by the emergence of new monitoring technologies.
The aim of such research is laudable: “The primary purpose of the exposome should be to identify risk factors in epidemiological studies. The aim is to generalize observations from a group of individuals to the population as a basis for public health decisions.”
Many projects are now pursuing this aim, especially in the field of cancer research, allergy research, early life exposures and children’s health and so on. Some can be found here on the website of the ‘Human Exposome Project‘. (In addition to the human exposome, there is also talk of an eco-exposome or an inter-exposome etc……, but this post is already too long)
Completeness, complexity, perplexity
While ever more sophisticated technologies might promise completeness, it has also become increasingly clear that the things they measure are very complex. The exposome is huge, has many components, is dynamic and varies over time and space, and, on top of this, exposures are always mixed and variable, as well as cumulative. This means even with the best of technologies coupled with the best of big-data analysis tools, deriving therapeutic insights will be difficult.
An article on the exposome and epigenetics summarises this very well: “Three layers of the exposome have been proposed: ‘general external’ (including social capital, stress and psychology); ‘specific external’ (including chemicals, viruses, radiation, etc.); and ‘internal’ (including for example metabolism and gut microflora). In addition, there are at least three properties of the exposome: (a) it is based on a refinement of tools to measure exposures (including internal measurements in the body); (b) it involves a broad definition of ‘exposure’ or environment, including overarching concepts at a societal level; and (c) it involves a temporal component (i.e., exposure is analyzed in a life-course perspective). The conceptual and practical challenge is how the different layers (i.e., general, specific external, and internal) connect to each other in a causally meaningful sequence.”
Only once this Gordian knot of complexities has been cracked, can exposome research be translated into health policies. This will be challenging.
Exposome research and the social sciences
Despite these difficulties, or perhaps because of them, some people have been calling for interdisciplinary collaborations between natural and social scientists. This is of course not surprising, as the exposome comprises not only chemicals but also capital, not only life but also lifestyle, not only pollution but also politics…
Some exposome researchers also seek links with experts in environmental justice and link exposome research historically to the beginning of the environmental movement and Rachel Carson’s Silent Spring. In France, these links are made explicit in an excellent overview article published by the University of Grenoble (worth reading; it also contains various useful illustrations, which are however copyright protected).
Similar calls for collaboration between natural and social scientists have been reverberating around epigenetics for a long time. I hope the the exposome doesn’t quite go the way of epigenetics which, as one of its lead researchers Edith Heard, has recently pointed out, has become: “une discipline en plein boom depuis le début des années 2000 et qui fait couler beaucoup d’encre de par les espoirs, mais aussi les fantasmes, qu’elle suscite” – some of these hopes and fantasies are flowing from the pens of social scientists.
The next big thing?
At the end of 2018 Veronique Greenwood published an article on exposome research entitled “The Next Big Thing in Health is Your Exposome”. It provides an overview of recent research by “Michael Snyder, a Stanford biologist and pioneer in genomics”. He talks about research he has carried out on himself using state-of-the-art monitoring devices, devices and technologies that have come a long way since 2005 and 2012!
He found that the exposome, his in this case, is vast and dynamic. However, he claims that “this is the first map of the human exposome, like the first genome map. We see what’s there, and then we try to understand how it affects your health. As we get more devices out, we will be able to make more associations between allergies and exposures. It would take long-term monitoring to understand the effects of toxins. But I do think we need that data.”
What do people think?? Is the exposome ‘the next big thing’, a new paradigm, a new frontier, or just a “wild idea“?
PS here is an overview article from 2019 that Aleksandra Stelmach just sent me
Image: Garbage, free image
No comments yet, fill out a comment to be the first

Leave a Reply