
September 15, 2017, by Brigitte Nerlich
Turning bacteria into passwords
A few days ago, I had the pleasure to meet two enthusiastic members of the first Nottingham iGEM team: Vikram Chhapwale, who specialises in computer science and AI, and Chris Graham, an emerging expert in biochemistry and genetics. Both are undergraduates studying at the University of Nottingham.
Chris and Vikram are part of a seven-member team which is made up of Jake, Ellie, Georgette, Matt, Natalia, Vikram and Chri. Together, they have backgrounds in biotechnology, biochemistry, genetics, chemical engineering and computer science – a great interdisciplinary mix. They get advice from international science, engineering and computer experts, such as Maria Zygouropoulou, Daphne Groothuis, Luca Rossoni, Sophie Vaud, Ines Canadas and Jamie Twycross, as well as from Louise Dynes, our outreach officer. You can find out more about their project here on the website of the Synthetic Biology Research Centre Nottingham, led by Nigel Minton, Alex Conradie and others.
I won’t provide more general background information on iGEM itself, as I have done that elsewhere. Let’s just say that iGEM is a student synthetic biology competition which aims to bring together young scientists from around the world, addressing global challenges through the engineering of biological parts.
Vikram, Chris and I had a great chat and I tried to learn about what they want to do for the upcoming iGEM competition. The operative word here is ‘tried’. Not being a maths, genetics and/or computer buff (rather the opposite), most of what they said went slightly over my head. In the following I’ll nevertheless try to give you a flavour of what the Nottingham iGEM project is all about – and in this case ‘all’ is the operative word. Entering the iGEM competition involves so much more than just ‘doing a science’ as the Sarcastic Rover would put it.
In the following, I’ll first talk about all that goes into ‘doing an iGEM’; then I’ll give you a flavour of Nottingham’s project and then I’ll say something about the personal journey on which the members of our team have embarked.
iGEM – life, the universe and everything
The undergrads involved in the iGEM competition have a lot of work to do over the summer, in preparation for the big ‘jamboree’ in Boston in November. They not only have to come up with a juicy science idea, they also have to produce a website, think about good ways of doing public engagement/human practices, get sponsors (in this case, for example, Eppendorff, Biolabs, SnapGene and, of course the University of Nottingham’s own Impact campaign), consult people on intellectual property issues, talk to potential investors and much more. As this is all happening for the first time for our team, this is a giant experiment, of which the little experiments in the lab are only a small part.
The team came up with a snappy title (“Key.coli”) for their project, which is looking at how biological systems can be used to overcome the limitations of current passwords; they set up a facebook page, a twitter account, a wiki, produced a Vlog, and created a catchy logo (which references Robin Hood – see featured image).
In order to stimulate public engagement with the project, they created two games that you can play and through which you can learn about the project. There is a 2D retro adventure game, inspired by current non-biological keys called THE KEY – which is also available on mobile and a game called FLUORESCENT ARCADE, both similar to Pac-Man games of the past; as well as a game called NOTTPORTAL, a 3D game inspired by the video game PORTAL, to be released at the jamboree in November.
iGEM – the key idea
When trying to come up with an idea for the scientific part of the iGEM competition, the team discussed the rise in cyberattacks and hacking which have recently been much in the news. In this context, ‘keys’, be they physical, digital and/or biometric, are of great importance and they have to be safe and secure. This poses problems, as physical keys can be ‘copied’ from a photo using 3D printing for example; digital codes can be hacked and ‘broken’ (and quantum computers are only just emerging as a remedy); fingerprints can be faked, and so on. So the team thought that it might be worth exploring the advantages (and possible disadvantages) of a ‘biosynthetic key’ or ‘password’ using bacteria, in this case E.coli.
The key idea for this new type of password that can be used to open accounts or safes is the use of a unique bacterium, indeed a bacterial password system (Key.coli). To achieve this, each bacterium is given a unique fluorescent property that is modularly created by a set of specific set of promoters and jellyfish fluorescent genes. In the key device, these bacteria are freeze-dried for storage and when the user needs to use it by “clicking” the metal case together, another vial filled with hydrating medium pours in, re-activating the bacteria and causes the key’s unique fluorescence to show. If this fluorescent spectrum matches (as decided by a similarity algorithm) the spectrum in the ‘lock’ or mother colony held within the scanning device, the safe or account can be opened. If the system is stolen, the bacteria die over the course of a week and the key becomes unusable.
This is the short version! The long version of the story involves proteins, plasmids, promoters, terminators, dead CRISPR Cas9, gene expression, ….. and quite a complicated way of ensuring randomness in the way genetic elements are assorted. So if you want to know more read on here.
iGEM – the journey
We all know that life is a journey. In the case of iGEM. the journey begins in Nottingham in the summer and ends in Boston in November (although ‘ends’ is the wrong word, as the students will probably carry iGEM with them throughout their whole life journeys). On the way, there is science to be done, which is a journey from an idea to an outcome and beyond, with lots of ups and downs; but there are also real journeys involved.
For example, the team went to a meeting in Edinburgh and enjoyed getting together up with other iGEM teams. They also went up the Shard in London, and so on. On the way they met lots of people and had all sorts of unexpected adventures. After an iGEM meeting in Nottingham, Vikram, for example, was invited to play the guitar at the Canalhouse Bar; Jake acquired skills in web design; Matt became a baker, and other members of the team have probably many more stories to tell.
iGEM – the excitement
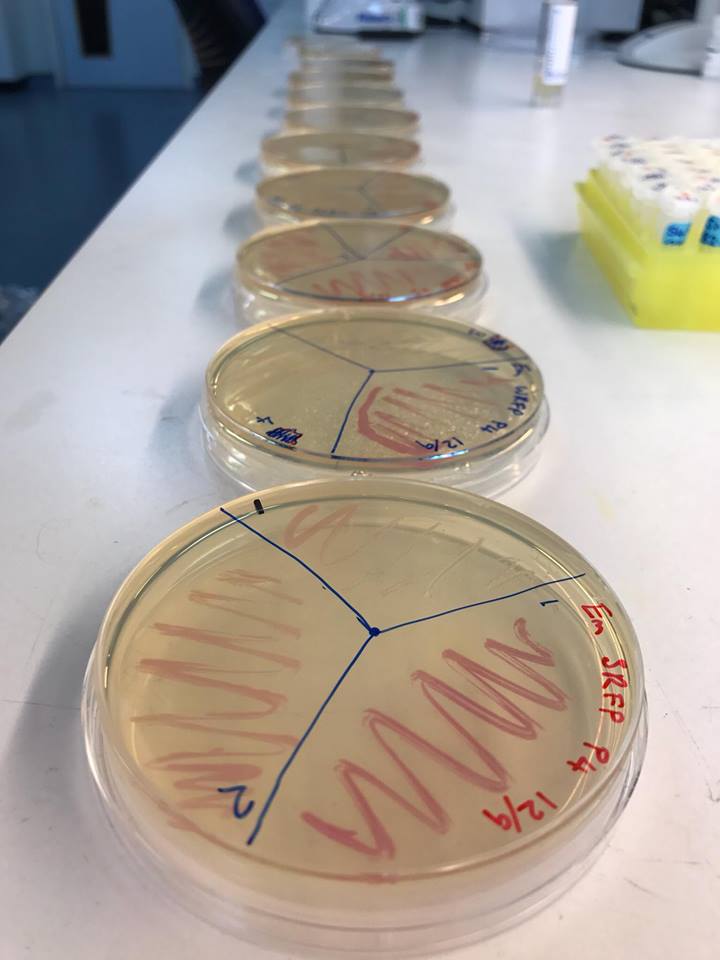
Working with bacteria is difficult. They don’t always do what you want them to do. But when they do, that’s very exciting. The day I was interviewing Chris and Vikram, Chris had just come out of the lab and found, what I heard in our conversation as ‘different shades of red’.
Here is Chris’s explanation of what that means: “When you spend every waking moment working with DNA, which can only be read from devices and screens, redesigning, retrying and theorising what you have designed will work, you can get nervous. This morning we saw the first colonies of our ‘Promoter Library’. They were all different shades of red and it meant that our modular design and use of promoters with alternate powers to express RFP [red fluorescent protein] was right. The fact we had a predictable array of red colour meant that we didn’t get everything wrong, the device is in our sights and finally… the cloning worked. I couldn’t be happier.”
I hope all goes well for the team and their bacteria in the autumn and I wish them all good luck inBoston, and lots of fun on the way!
No comments yet, fill out a comment to be the first

Leave a Reply